NeuroFeatureExtract
Overview
The EBRAINS NeuroFeatureExtract (NFE) is a web application that allows users to extract an ensemble of electrophysiological properties from voltage traces recorded upon electrical stimulation of neuronal cells (see Fig. 1). The main outcome of the application is the generation of two files - features.json and protocols.json - that can be used for later model parameter optimizations.
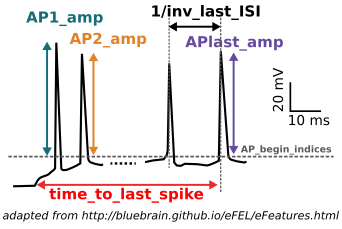
For data selection, there are three possible scenarios:
1) The user selects voltage traces from the provided dataset;
2) The user selects the voltage traces from uploaded data files (through the “Upload” utility of the
GUI);
3) The user selects traces
from both the dataset and her/his own uploaded file(s).
This application leverages the Electrophys Feature Extract Library (eFEL) and the
BluePyEfe
software packages and provides a friendly interfaceto select voltage traces (individually or
collectively,
based on the applied stimulus
current) and features to be extracted.
Electrophysiological data
The NFE can only be used for voltage traces recorded or simulated during a step current stimulation protocol. The files uploaded by the users must fulfil this requirement. Below we report a brief description of the data formats currently in use.
Dataset
Publicly available data consist of electrophysiological traces provided by contributing laboratories internal and external to the Human Brain Project (see Fig. 2).
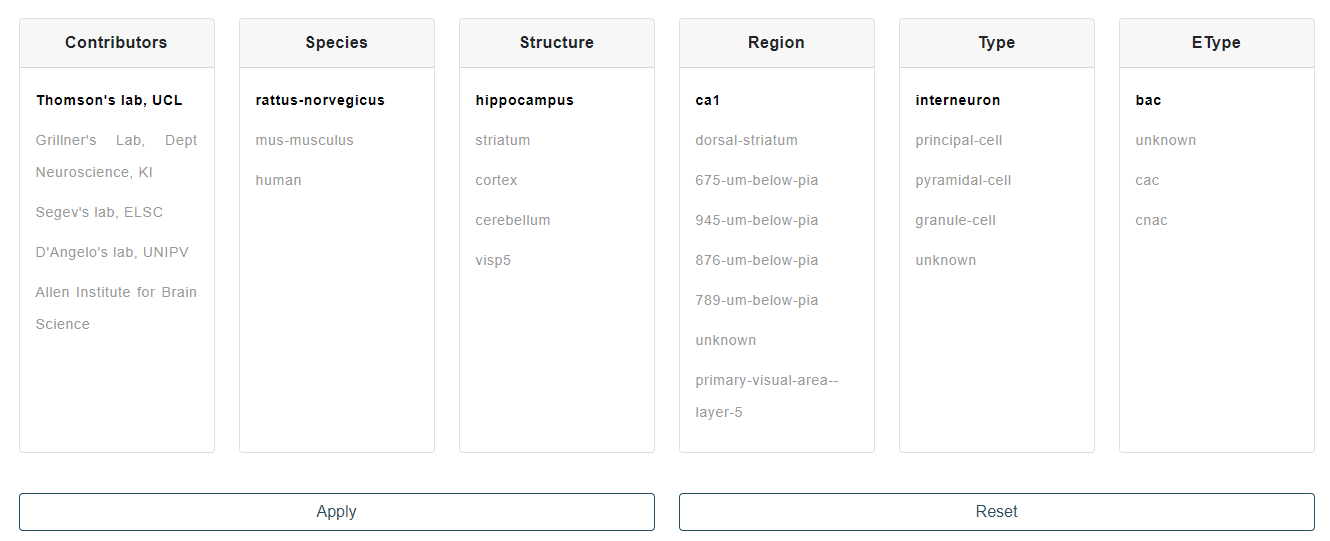
Contributed raw data files are in the Axon Binary Format (.abf) or the Igor Binary Format (.ibw) and have been converted to .json files and stored in a dedicated CSCS Object Storage container after insertion of relevant metadata (e.g. cell_id, brain_region, brain_structure, stimulus_duration, …) used for data processing and visualization. The data are read through the Neo and Igorpy Python packages and converted into value arrays before use. If you would like to contribute with your own data to the NFE, please send an email to support AT ebrains.eu.
User’s uploaded data
Users can also upload and visualize their own data, which can be selected, for processing, independently
or
together with traces from the provided dataset.
For detailed information on the accepted data formats, please visit the NFE Guidebook dedicated
page.
Feature Extraction
The feature extraction process precedes the generation of the features.json and
protocols.json
files, which are used for model parameter optimization performed through the BluePyOpt software tool (please refer to the BluePyOpt documentation for detailed explanation) or for statistical
analysis.
The NFE is integrated in the
Hodgkin-Huxley Neuron Builder web application that provides a point-and-click user
friendly interface to guide the user through the entire optimization process.
The features that the user can select for extraction are described in the eFEL documentation as well as
the eFEL software package
code. See also the Hippocampal Neurons dedicated page in the
EBRAINS Cellular Level
Simulation Interactive Workflows and Use Cases guidebook.
Features are computed for every trace of the chosen recordings, where a trace corresponds to a given
stimulus current amplitude (indicated to the user when data are displayed). Once the extraction is
finalized,
the values of a feature obtained from traces belonging to an individual cell and corresponding to the
same
stimulation amplitude are averaged. The averages computed for all the cells, are averaged a second time
by
stimulus amplitude. This will generate two result files (i.e. features.json and
protocols.json) per cell
plus
two supplementary files with the global averages.
Graphical User Interface
The GUI guides the user through the feature extraction process in a friendly way. The web application
homepage shows a short tutorial on the usage of the interface.
The following is a description of the Feature Extraction process.
Trace Selection
Once the trace selection page is displayed, data can be filtered by choosing the following properties of
the
cell from five menu lists: 1) Contributor; 2) Species; 3) Brain structure; 4) Region; 5) Type; 6)
Electrical
type (eType). If any of the fields is missing (e.g. the eType of the
cell
is not known), the “unknown” label
is displayed.
When the data are loaded, the traces contained in each file are shown and data files are grouped by cell
id
(see figure below). The user can select individual traces by clicking on the corresponding amplitude or
(s)he can select/deselect all traces in a single file. Additionally, all files (and then all traces)
referring to a single cell can be selected (see Fig. 3).
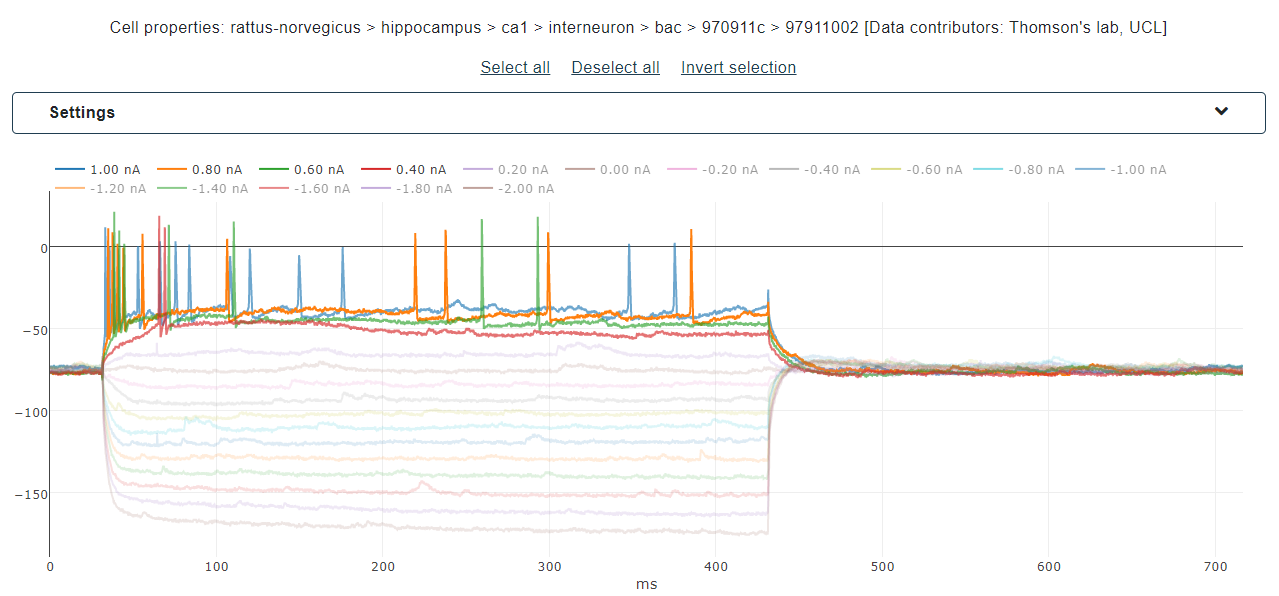
Alternatively, or concurrently, users can upload their own data (see User’s uploaded data) by browsing their local storage and using the upload button (see figure below). The traces will be displayed for selection, together with the ones selected from the HBP dataset (if any).
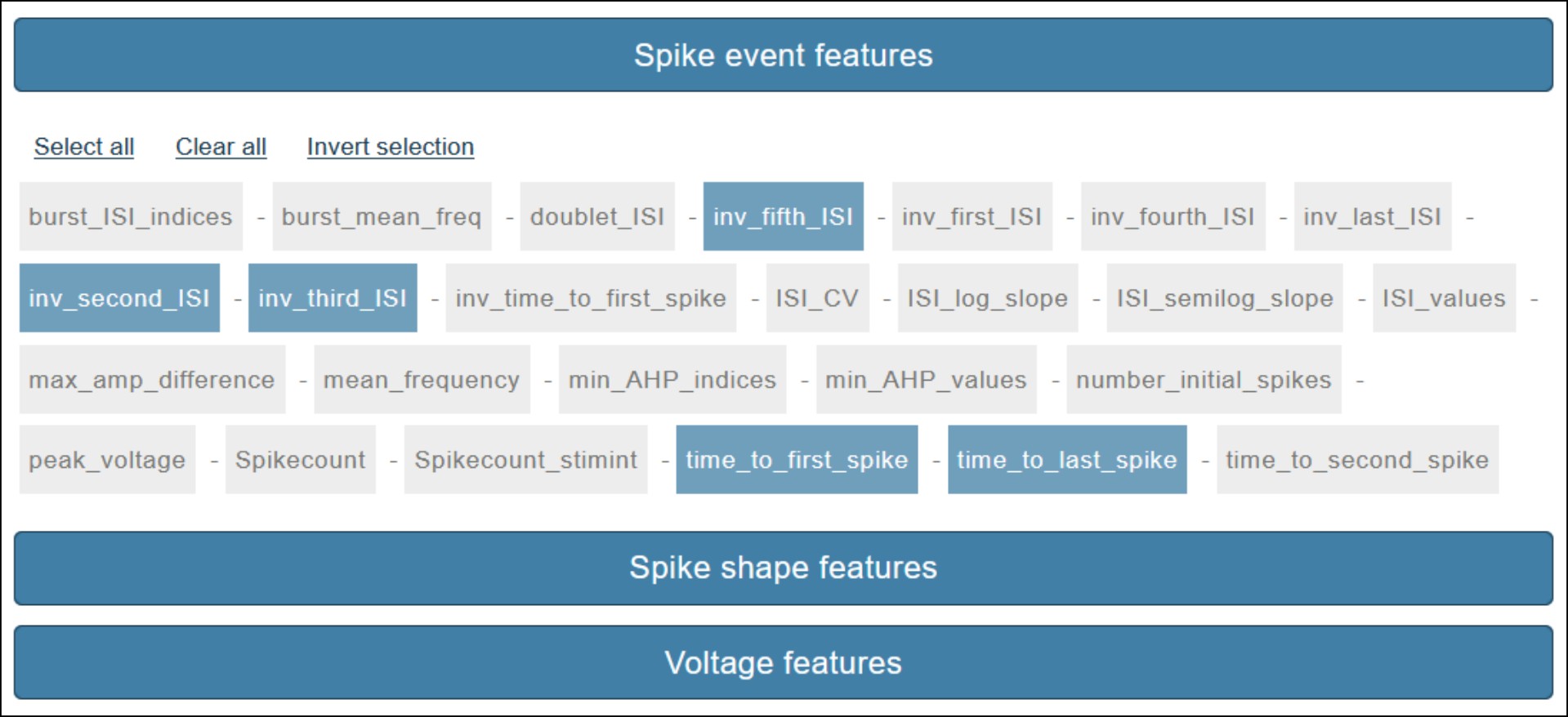
Feature selection
Once the trace selection approved, the feature selection page is displayed. Features are grouped by type -spike event features, spike shape features, voltage features- and can be selected individually or collectively through the select/deselect buttons. Given the high number of features, the three types are grouped in toggle boxes (see Fig. 4). Upon selection approval, the feature extraction process takes place
Results
Finally, a success message is displayed and results are made available to download. The application output consists of a features.json and protocols.json files which are generated for both individual cells and the entire ensemble. These files contain the feature value averages (computed as outlined in the above section “Feature Extraction”) and the protocols adopted for the experimental recordings. These files are intended to be used for the data-driven optimization step of the Hodgkin-Huxley Neuron Builder workflow.
Contacts
We are always happy to hear from users and to get feedback to improve our tools and services. Drop us an email with your feedback or ask your questions by filling out the dedicated form.
