The NeuroFeatureExtract tool allows users to select data from an HBP dataset and/or upload their own, and extract electrophysiological features of interest.
The application leverages the Python Electrophys Feature Extract Library (eFEL) and provides a friendly interface to select both individual voltage traces (based on the stimulus current applied) and features to be extracted.
Please find below a short tutorial on how to use the application or read the
guidebook to get started
and get the references to cite our work.
Select the data from a dataset, based on cell properties to be chosen from the filter dropdown menus. Additionally and/or alternatively upload your own data for processing (see Fig. 1). The only file extension allowed at present is .abf (axon binary file) and the uploaded files must contain both the voltage and the stimulus signals. This is a mandatory condition for several features to be computed and for the feature extraction process to be completed successfully. The analog voltage and stimulus signals are extracted through the neo python library, designed for representing different formats of intracellular and extracellular electrophysiological data (see neo for a complete reference).
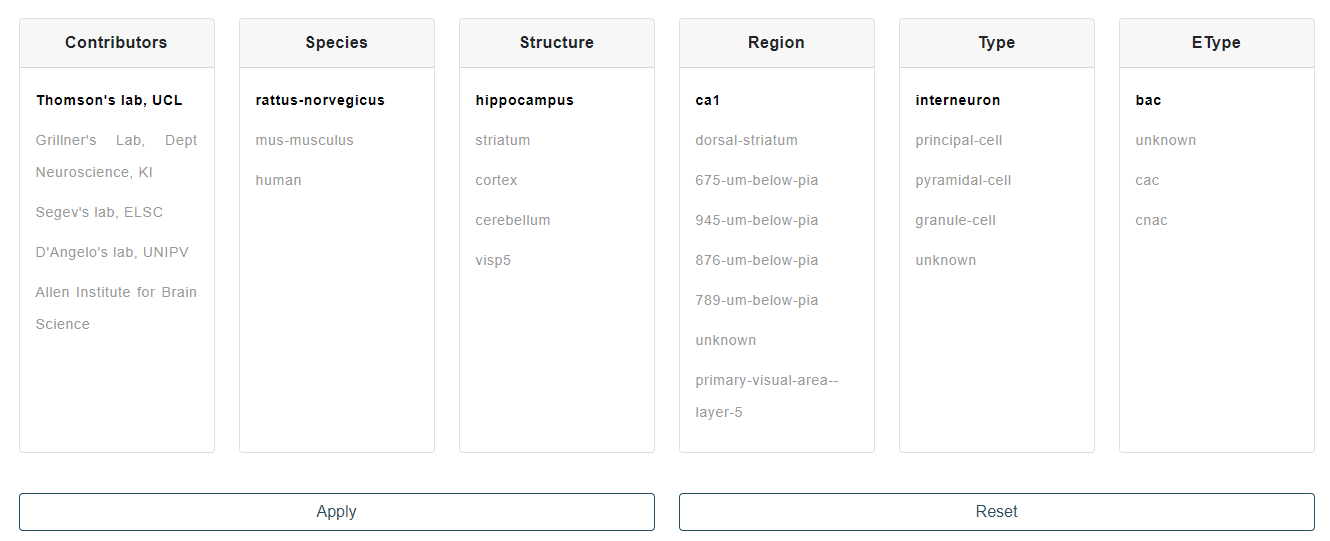
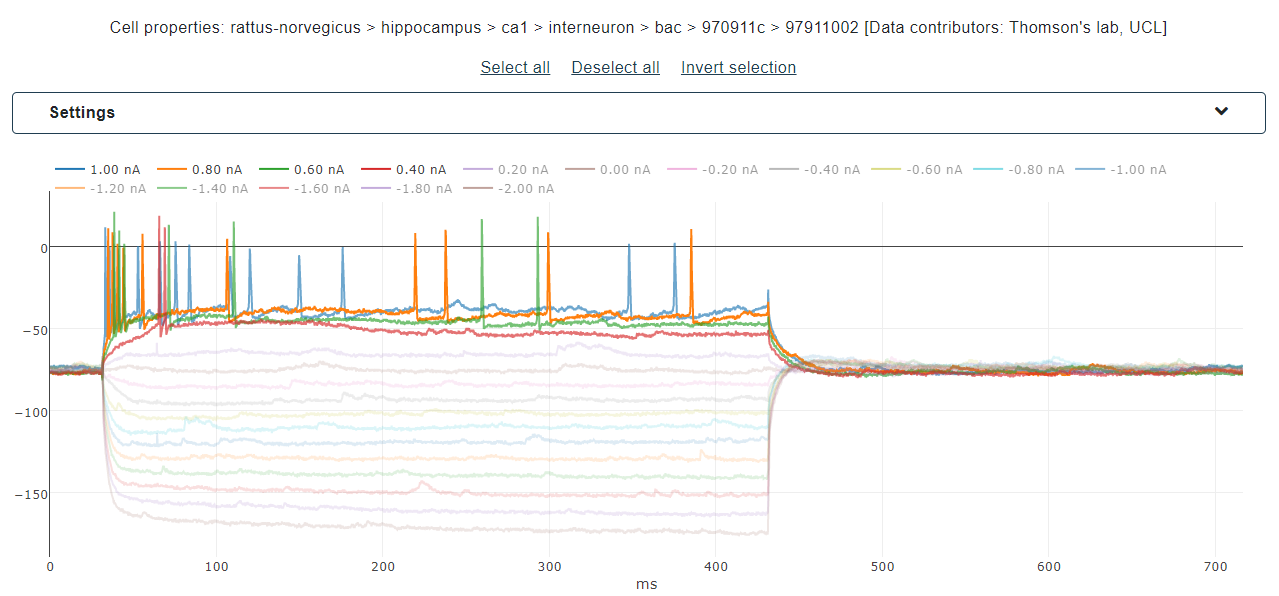
Once the filtering/upload done, individual traces are plotted and can be checked for selection. Traces represent the voltage membrane responses of the selected cell to the stimulus displayed in the legend. Highlight individual traces by hovering on the corresponding stimulus and select them by clicking on the same legend or through the selection buttons (see Fig. 2). When all the traces of interest have been selected, go the next page for feature selection.
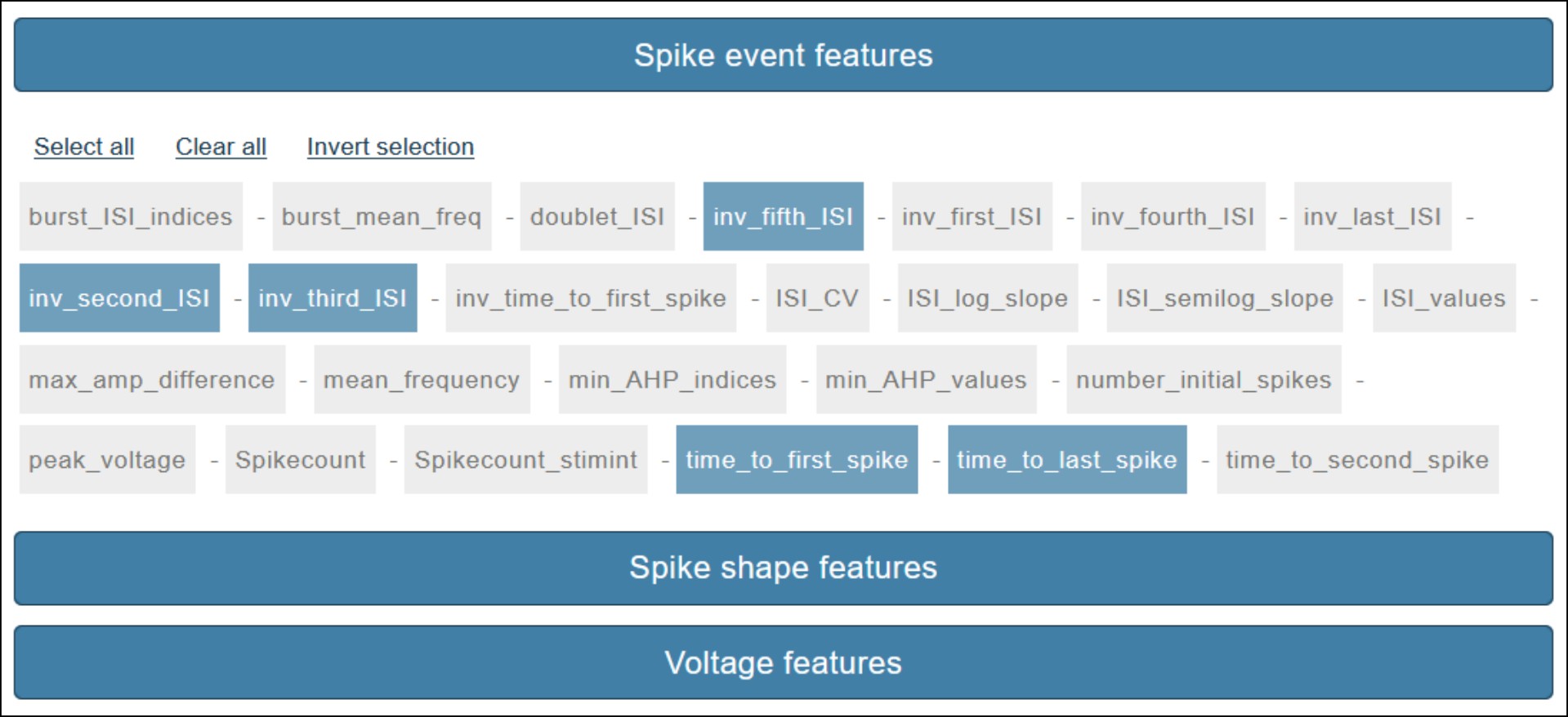
The feature selection page allows you to select the features to extract from the electrophysiological chosen signals. Features are grouped by type: 1) Spike event features - 2) Spike shape features and 3) Voltage features, and are selected by clicking on the corresponding box. When hovering on feature names a brief description is provided (see Fig. 3). Once the selection is completed, click on the Next button to launch the extraction process.
The feature extraction process computes the means and standard deviations of the
selected
features for individual cells and for individual stimuli. Once a stimulus is chosen
for a single
file, it will be taken into account for all the selected files corresponding
to the same cell. Finally, a grand mean is computed among different cells.
A features.json and protocols.json files are generated for both
individual cells and
the entire ensemble. These files contain the above mentioned grand-means and the
protocols adopted for the experimental
recordings.
They are intended to be used for the data-driven optimization step of the
Hodgkin-Huxley Neuron Builder workflow
(via the
BluePyOpt
software library), made available to the user at this
link.
A link to a .zip file containing the entire set of results is provided in the
results page (see Fig. 4).
